 +565 975 658
+565 975 658
 info@premiumcoding.com
info@premiumcoding.com
 Monday - Friday, 8.00 - 20.00
Monday - Friday, 8.00 - 20.00
As is known from interdisciplinary research about biomaterials, chemical biology, cell biology and regenerative medicine, biomaterial surfaces can lead to various cell responses. Recently, Ding’s group has revealed that well designed surface topography can even induce severe nuclear deformation, chromosomal repositioning and changes of gene expression.
With a silicon micropit template prepared with microelectronics manufacturing technology, Ding’s group prepared micropillar arrays of PLGA, an important polymeric material. Under an appropriate micropillar array, cell nuclei exhibited significant self deformation, as shown in Figure 1.
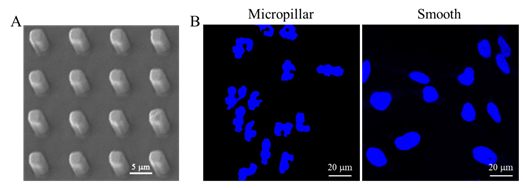
Figure 1. Cell nuclear deformation on a PLGA micropillar array. (A) SEM image of the PLGA micropillar array. (B) Fluorescence images of the nuclei of HeLa cells on the PLGA micropillar array and smooth surface.
Ding’s group further elicited two questions: will the positioning of some chromosomes change on the micropillar array in comparison with those on the smooth surface, and will gene expressions of the cells vary on the topological surfaces along with nuclear deformation? To this end, fluorescence in situ hybridization (FISH) was employed to detect the radial disposition of chromosomes 18 and 19 (Figure 2). These chromosomes were selected as two models, because chromosomes 18 and 19 have similar DNA contents but contrasting gene densities, and are typical for examination of chromosomal territory. It was indicated that chromosomes 18 and 19 moved towards the nuclear periphery in response to the cell nuclear deformation on the micropillar array, with chromosome 18 translocated much closer to the cell nuclear periphery.
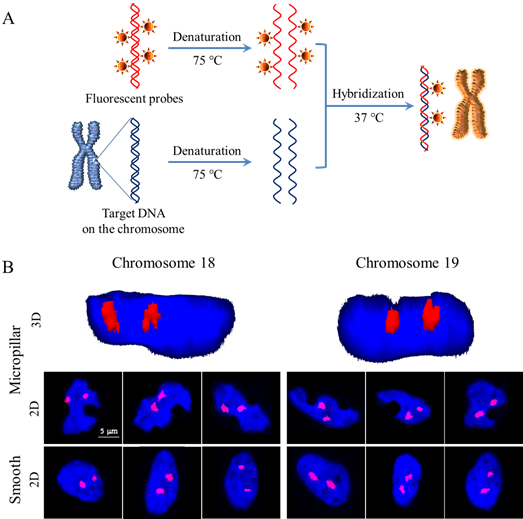
Figure 2. FISH staining for monitoring chromosomal territories of chromosomes 18 and 19 of HeLa cells on the micropillar array and smooth surface. (A) Principle of fluorescence in situ hybridization (FISH). (B) 3D and 2D fluorescence images of chromosomes 18 and 19. The chromosomes were hybridized with whole-chromosome painting probes with orange fluorochrome, and the nuclei were stained blue with DAPI.
DNA microarray technology was also used to detect the gene expression profiles of the cells on the PLGA micropillar array (Figure 3). It was found that 180 genes were up-regulated and 255 genes down-regulated for the cells on the micropillar array. The corresponding transcriptome was further investigated via Bioinformatics using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses.
The article was published in the journal of ACS Applied Materials & Interfaces in August, 2020. The first author is Dr. Ruili LIU and the corresponding author is Prof. Jiandong DING.
Publication link: https://dx.doi.org/10.1021/acsami.0c05883
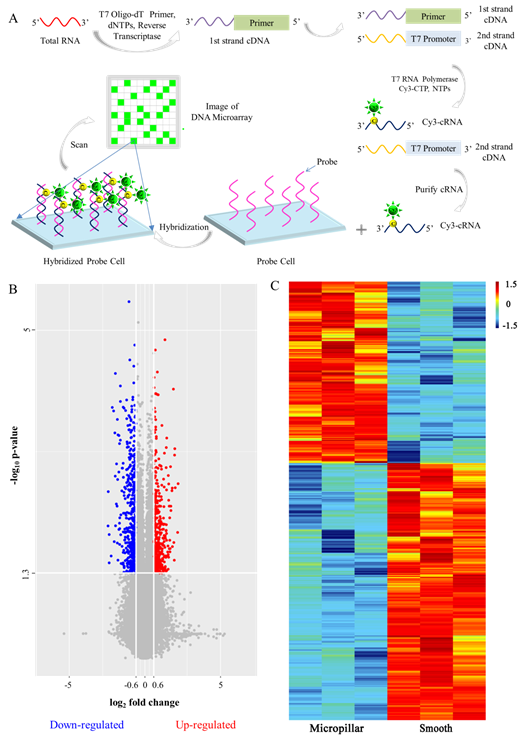
Figure 3. Gene expression of all of the chromosomes in HeLa cells detected with a DNA microarray. (A) Process of the detection of total RNA with a DNA microarray. (B) Volcano plot of differentially expressed genes of HeLa cells on the micropillar array. For each group, n = 3. The data with p-value of fold change lower than 0.05 (-log10 p-value higher than 1.3), and fold change lower than 1/1.5 (log2 fold change < -0.6) indicates the down-regulation of the gene expression of HeLa cells on the micropillar array, which is shown in blue. The data with p-value of fold change lower than 0.05 (-log10 p-value higher than 1.3), fold change higher than 1.5 (log2 fold change > 0.6) indicates the up-regulation of the gene expression of HeLa cells on the micropillar array, which is shown in red. (C) Heatmap of the differentially expressed genes of HeLa cells on the micropillar array (fold change > 1.5, p < 0.05). Red, high expression; Blue, low expression. The values of 1.5, 0 -1.5 in the ruler reflect the maximum, medium, and minimum values of gene signals of the differentially expressed genes. The rows reflect the samples, and every line reflects one gene.
It is interesting that an appropriate micropillar array can trigger significant self deformation of cell nuclei on the subcellular level. Besides the change of the cell nuclear shape, such a special topological cue of a material can lead to repositioning of chromosomes on the sub-nuclear level. The topological morphology of the biomaterial can further induce a large amount of genes to be up or down regulated. The surface-topography-induced chromosome repositioning and corresponding significant regulation of gene expression should be considered in biomaterial design for, in particular, cancer treatment, tissue engineering, and regenerative medicine.
This work was published on ACS Applied Materials & Interfaces with Dr. Ruili LIU as the first author. See please: Ruili Liu, Jiandong Ding*, Chromosomal repositioning and gene regulation of cells on a micropillar array. ACS Appl. Mater. Interfaces, 12, 32: 35799 - 35812 (2020).
Article links: https://dx.doi.org/10.1021/acsami.0c05883
Get to know us better now!

Wechat:FDUMMers
Search!
Search across our website
Revenant @ 2018 by fudan | All Rights Reserved
Powered by Weicheng

