 +565 975 658
+565 975 658
 info@premiumcoding.com
info@premiumcoding.com
 Monday - Friday, 8.00 - 20.00
Monday - Friday, 8.00 - 20.00
Cell migration is regulated by RGD nanospacing and enhanced under moderate cell adhesion on biomaterials
Cell migration on a biomaterial is critical for tissue engineering, tumor metastasis, and regenerative medicine, in particular, for in situ tissue induction after implanting biomaterial scaffolds with bioactive internal surfaces. Ding’s group has developed a surface patterning technique to investigate the effects of ligand nanospacing on cell adhesion. Recently, the group further revealed that cell migration was enhanced under appropriate nanospacing with moderate cell adhesion on biomaterials.
Block copolymer micelle nanolithography was used to prepare gold nanopatterns as shown in Figure 1. Further forming self-assembly monolayers of RGD peptide led to RGD nanopatterns. Since RGD is a ligand in extracellular matrix to bioconjugate with integrin, its receptor in the cell membrane, the series of RGD nanopatterns with a nonfouling background enabled the researchers to manipulate the integrin distribution and thus cell adhesion and migration.
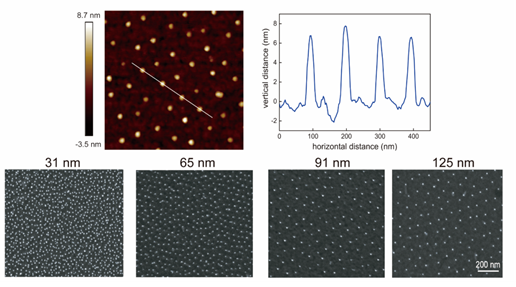
Figure 1. AFM and SEM images of as-fabricated nanopatterns.
Ding’s group examined the adhesion and motility of human umbilical vein endothelial cells (HUVECs) on the well-controlled RGD nanopatterns. The endothelial cells exhibited a higher cell density and larger cell spreading area on nanopatterns of smaller RGD nanospacings, consistent with the previous observations of various other types of cells on RGD-nanopatterned surfaces. Cell migration exhibited a nonmonotonic change with the ligand nanospacing: the maximum migration velocity appeared around 90 nm of nanospacing (even much faster than the group of “non-nano”), and slow migration happened in the cases of small or large RGD nanospacings. Therefore, moderate cell adhesion is beneficial for fast cell migration. Both the single cell migration and the collective cell migration assays showed the same results (Figure 2).
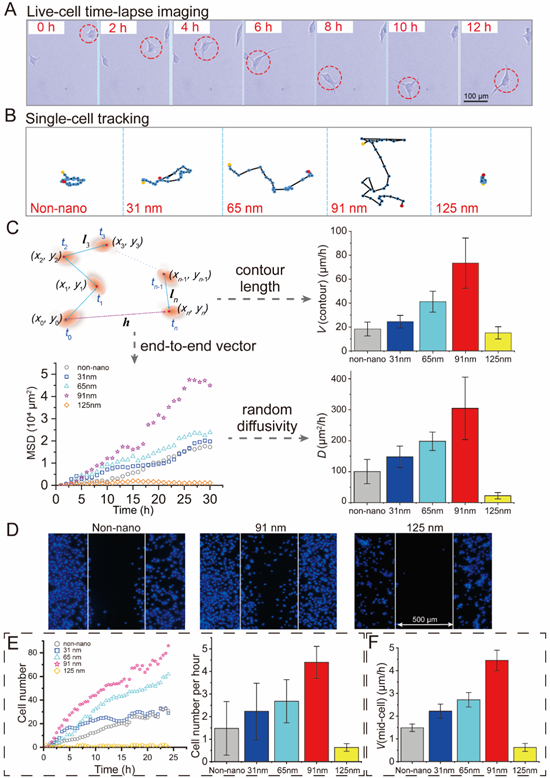
Figure 2. Cell migration on RGD-nanopatterned surfaces. (A) Live-cell time-lapse imaging of HUVECs at a low seeding density to show single-cell migration on a nanopattern with 91 nm of nanospacing. (B) The corresponding trajectories of single endothelial cells on the nanopatterned surfaces with the indicated RGD nanospacings, and on the non-nano gold surface treated by RGD-thiol ligands as well. Each site represents a time-dependent position of the center of mass of a single cell. (C) Statistics of the tracking pathways of single cells with the random-walk model. Both the calculated V(contour) and D showed nonmonotonic trends for cell migration on RGD-nanopatterned substrates, and a peak was found around 91 nm of nanospacing. (D) Representative images of collective cell migration on the indicated substrates after culturing for 24 h using wound healing assays. The white lines indicated the original gap produced by the culture inserts. Cells on 91 nm-nanospaced substrates migrated faster to fill the gap than all of the other groups. (E) Counting of the numbers of cells migrated into the “scratch” areas along with the record time (left) and the calculated migrated cell numbers per hour reflecting the quantity of the collective migration (right). (F) Calculated V(mid-cell) reflecting the speed of the collective migration. The velocities were calculated on the basis of the distances between the middle cells and the initial border lines at 24 h.
In order to interpret this phenomenon in molecular level, some key genes and proteins related to dynamic actin rearrangement were analyzed, and the maximal cell migration corresponded to the upregulation of activated small G-proteins, such as Cdc42, Rac1 and RhoA (Figure 3).
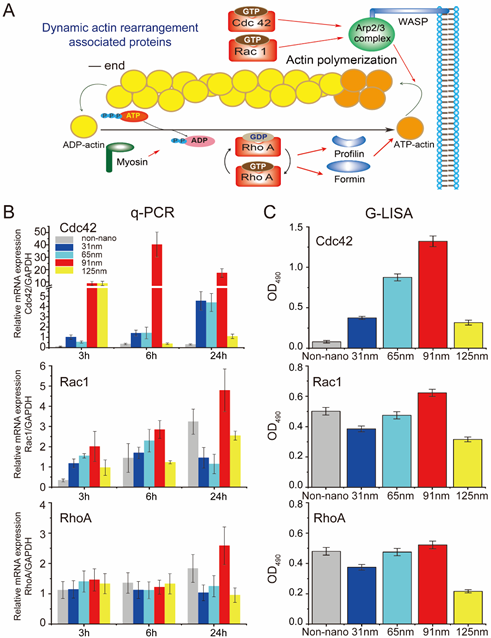
Figure 3. q-PCR and G-LISA results for some selected key genes and proteins related to dynamic actin rearrangement of cells. (A) Schematic illustration of the proteins associated with dynamic actin rearrangement of cells, and the molecular mechanism of the regulation of those proteins on actin dynamics. (B) mRNA expressions of the genes related to dynamic actin rearrangement quantified by q-PCR showed nonmonotonic trends with peaks at 91 nm of RGD nanospacing. (C) G-LISA results presented as the optical densities at 490 nm. The active GTP-bound levels of the Rho proteins, which are tightly associated with dynamic actin rearrangement, also showed nonmonotonic trends with peaks at 91 nm of RGD nanospacing.
Overall, this study revealed that modification of a biomaterial with compromised cell adhesion may achieve superior cell migration; an appropriate nanoscale modification can significantly enhance cell migration, while some other nanoscale modifications might lead to the opposite. These findings are potentially very helpful for guiding the design of biomaterials for promoting tissue regeneration with enhanced cell migration or inhibiting tumor metastasis with depressed cell migration.
The article was published in the journal of Biomaterials with Dr. Qiong Liu and Ms. Shuang Zheng as co-first authors and Professor Jiandong Ding as corresponding author. See please: Qiong Liu#, Shuang Zheng#, Kai Ye, Junhao He, Yang Shen, Shuquan Cui, Jiale Huang, Yexin Gu, Jiandong Ding*, Cell migration regulated by RGD nanospacing and enhanced under moderate cell adhesion on biomaterials, Biomaterials 263, art. No. 120327 (2020).
Article link: https://www.sciencedirect.com/science/article/pii/S0142961220305731
Get to know us better now!

Wechat:FDUMMers
Search!
Search across our website
Revenant @ 2018 by fudan | All Rights Reserved
Powered by Weicheng

