 +565 975 658
+565 975 658
 info@premiumcoding.com
info@premiumcoding.com
 Monday - Friday, 8.00 - 20.00
Monday - Friday, 8.00 - 20.00
Cell adhesion to extracellular matrices (ECM) is critical to physiological and pathological processes as well as biomedical and biotechnological applications. To reveal the cell adhesion rules on the surface of ECM-like biomaterials, especially the relevant regulation of cell behavior, nanocue, is of fundamental significance for tissue engineering, regenerative medicine and tumor targeting. It’s also one of the keys to guide the design of new materials for biomedical and biotechnology applications.
Ding’s group has developed a unique micro-nanopatterning technique to investigate the critical adhesion areas of cells (Figure 1), and investigated the relationship between critical area and nanospacing of ligands fixed to the surface of the material. They further speculated the minimal effective RGD (arginine-glycine-aspartate) nanospacing on unpatterned free surfaces with densely grafted RGD.
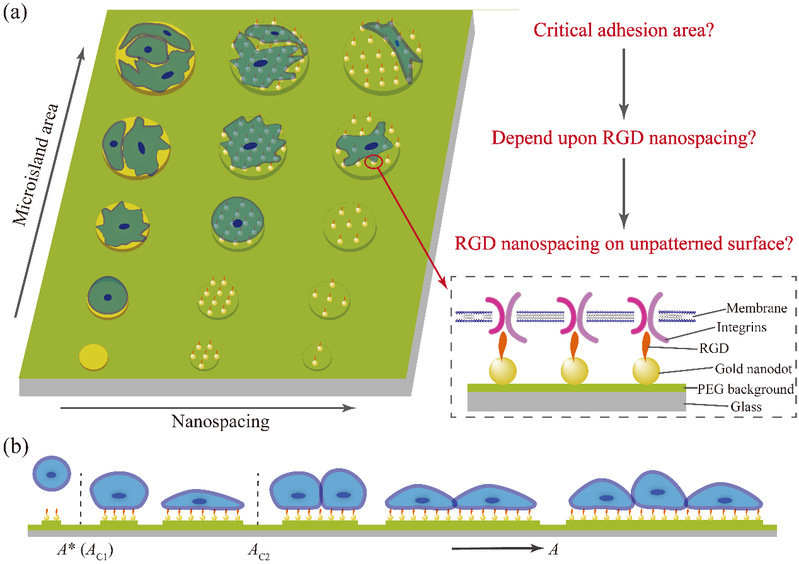
Figure 1 Schematic illustrations of the basic scientific questions and key terms. (a) Micro-nanopatterns and micropatterns of tunable microisland sizes and RGD nanospacings to control cell adhesion; (b) critical adhesion areas of individual cells on adhesive nanoarrayed microislands with nonfouling background.
Unlike individual micropattern or nanopattern, the micro-nanopattern prepared by Ding's group can control both the sizes of the microislands and the spacing between the gold nanodots at the same time.
The main stages to fabricate the micro-nanopatterns are schematically represented in Figure 2, where they combined block copolymer micelle nanolithography for nanopatterning, photolithography for micropatterning, and hydrofluoric acid (HF) etching. Then, an anti-specific adhesion background was obtained by salinization. Finally, arginine-glycine-aspartic acid (RGD) polypeptide was linked to prepare a series of RGD micro-nanopatterns with cell adhesion contrast (Figure 2 & 3).
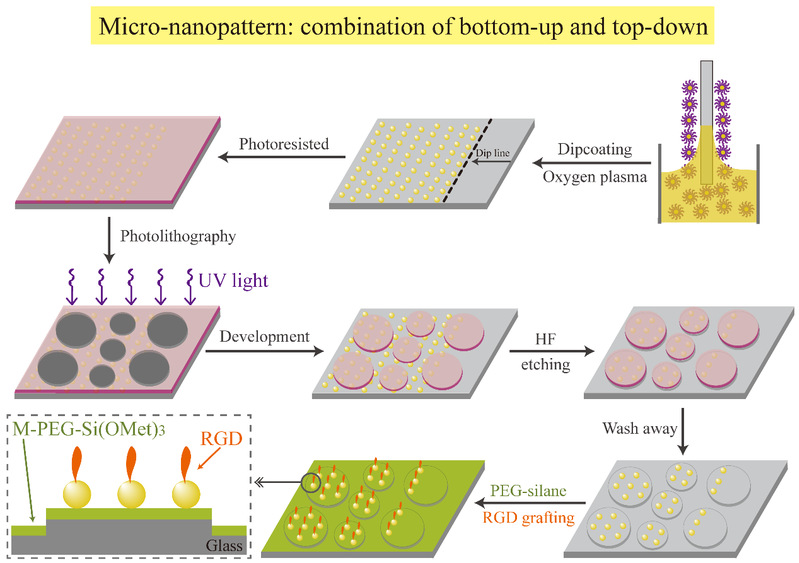
Figure 2 The main steps to fabricate the micro-nanopatterns.
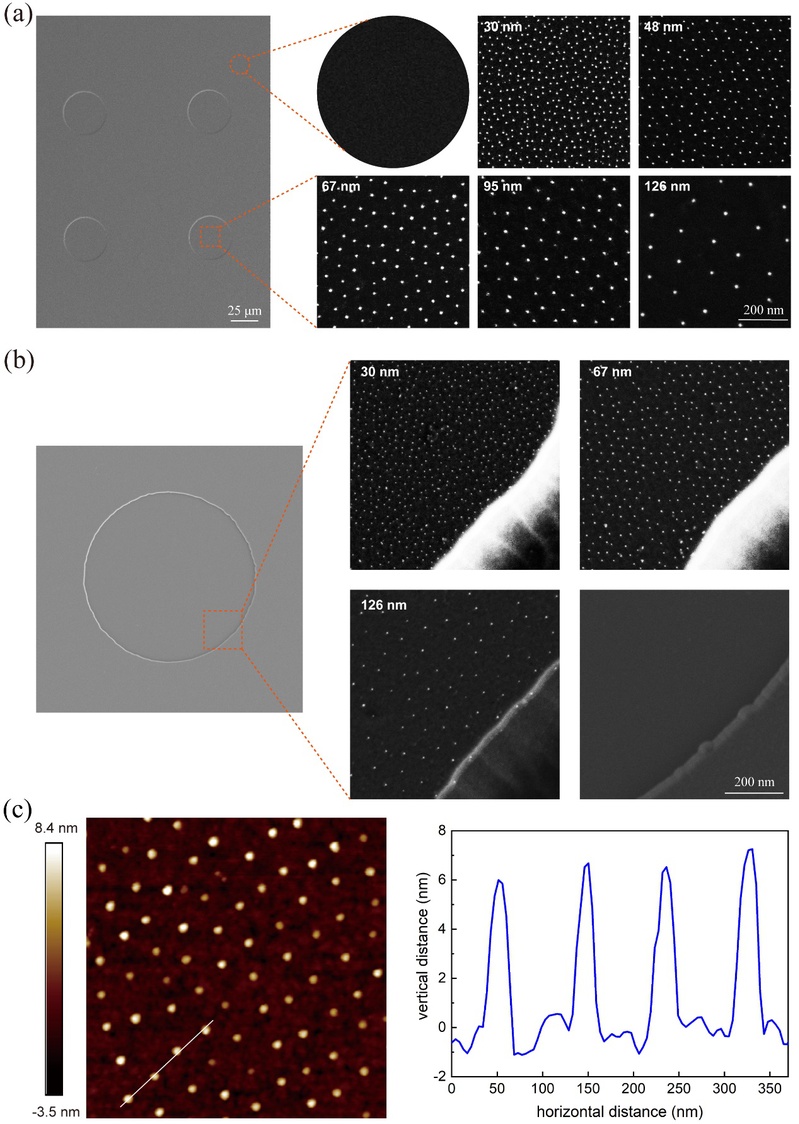
Figure 3 (a) FE-SEM images showing gold nanodots within the microislands (square images in the right column) and the background region outside the gold-nanodot arrayed microislands (round image in the right column); (b) FE-SEM images of a representative microisland (left column), the edges of gold-nanodot arrayed microislands of three micro-nanopatterns corresponding to three different nanospacings (right column), as well as the edge of an unnanoarrayed microisland on a micropattern as control; (c) Microscopic observations of nanopatterns by AFM.
Arginine-glycine-aspartate (RGD) is a kind of ligand in ECM which can bind to the receptor Integrin on the cell membrane. The micro-nanoarray of RGD can regulate the spatial distribution of integrin on the cell membrane, thus regulating the specific adhesion of cells and related cell behavior.
Ding’s group used this technique to investigated the number of human mesenchymal stem cells (hMSC) (from zero cell to several cells) on a series of micro-nanopatterns with different RGD nanospacings (30 - 126 nm) and microisland sizes (10 - 80 μm), and found that there exists a critical cell adhesion area on the micro-nanopatterns. Moreover, the critical adhesion areas increase with the increasing of RGD nanospacing (Figure 4).
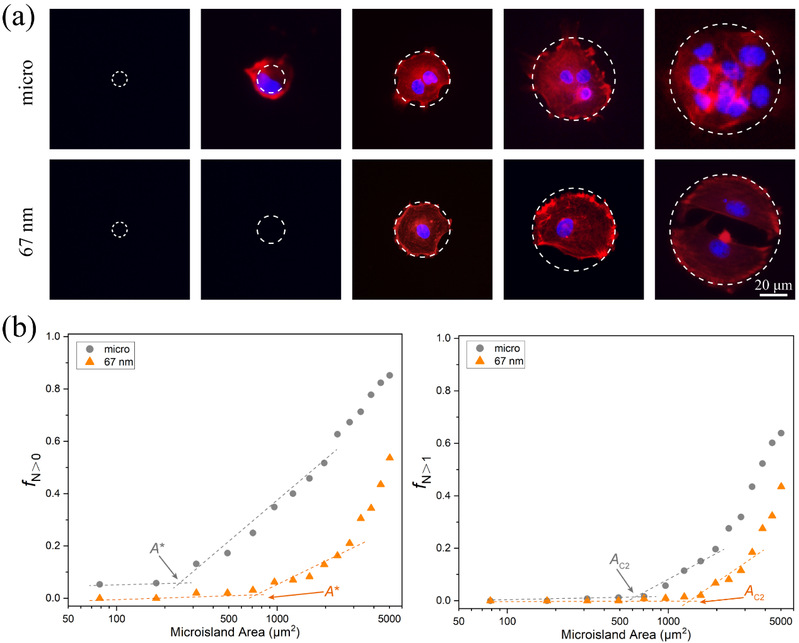
Figure 4 Critical adhesion areas of cells obtained by observation of cell numbers on microislands with increased sizes. (a) The relationship between cell numbers and the size of microislands together with nanospacings of micro-nanoislands. The F-actins were stained in red, and the nuclei were stained in blue. The number of nuclei indicate that of adherent cells on each micro/micro-nanoisland within the contour marked by the dashed circles. (b) The experimental approach to determine critical adhesion areas of cells on micro-nanopatterns with increasing microisland sizes. Left: fraction of cellular occupation per microisland (fN>0) on mere micropatterns (gray circles) and on 67 nm nanospaced micro-nanopatterns (orange triangles). Right: fraction of occupation of multiple cells per microisland (fN>1) on micropatterns and on 67 nm nanospaced micro-nanopatterns. The critical areas A* and Ac2 are from the crosspoints between the corresponding two marked asymptote lines of f N>0 and f N>1, respectively.
The significance of this fundamental research is not limited to the cellular behavior of patterned surfaces.Thanks to the development of nanopatterning techniques, it has been recognized that RGD nanospacing is critical for specific cell adhesion, and cells can adhere well only on a surface with a relatively small RGD nanospacing. Nevertheless, it does not mean that extremely small nanospacing is necessary for excellent cell adhesion. Even an unpatterned free surface with very dense RGD grafting might not correspond to a “zero” RGD nanospacing, not only because the sizes of RGD ligands for steric hindrance but also because the effective lateral distance of such ligands for collectively transducing signals from outside to inside of a cell. However, no one has revealed the exact effective RGD nanospacing for an unpatterned surface with a dense self-assembled monolayer of RGD ligands. Herein, Ding’s group discovered this “hidden” value assisted by the micro-nanopatterning techniques and the corresponding analysis based on the concept of critical cell adhesion areas.
The calculations of the first critical adhesion area (A*) (a single cell from non-adhesion to adhesion on micro-nanoislands) and the second critical adhesion area (AC2) (cells from single cell adhesion to multi-cell adhesion on micro-nanoislands) results in the almost consistent minimum effective nanospacing of the ligands, which were about 19 nm (Figure 5).
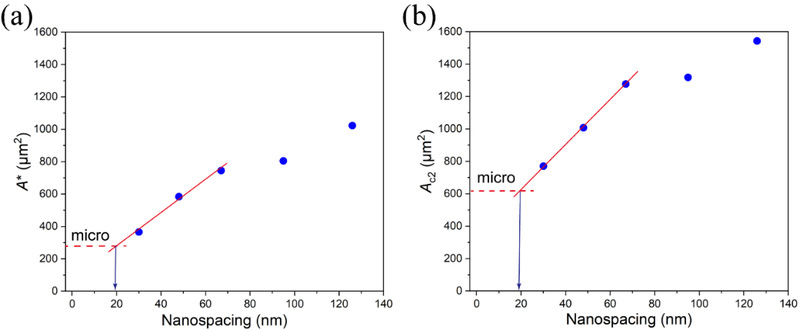
Figure 5 Dependence of the critical adhesion area on RGD nanospacing on a series of micro-nanopatterns and the asymptotic analysis of the effective RGD nanospacing on a free surface without patterns. A*(a) and AC2(b) increased with RGD nanospacing. The partial linear fitting curves for nanospacing < 70 nm of A* and AC2 were used to deduce the effective RGD nanospacing of an unpatterned surface with densely grafted RGD ligands, resulting in A* » 19.14 nm and AC2» 19.12 nm.
Based on the characteristic of micro-nanopatterns, the author further calculated the critical number of integrin-ligand binding (N*) by using the experimental results. The results showed that N* tended to decrease with the increasing of RGD nanospacing (Figure 6), and the number was not a constant.
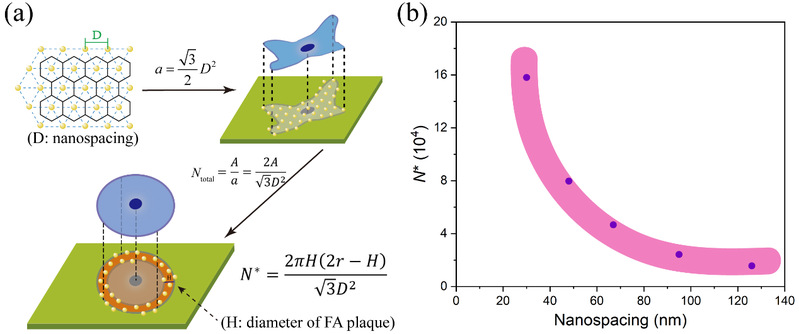
Figure 6 (a) Schematic illustration to demonstrate the deduction process of the possible critical number of integrin-ligand binding (N*); (b) The curve of N* versus nanospacing based on experimental results.
Based on the data treatment method of statistical physics to examine the finite size effect, the author proposed and deduced the background adhesion of cells on the material surface. Together with the specific cell adhesion, background adhesion provides the adhesion force of cells on the material surface, and the total adhesion force of cells under the critical adhesion area is a conserved quantity (Figure 7).Eventually Ding’s group put forward a unified interpretation by combining nonspecific background adhesion and specific cell adhesion.

Figure 7 Derivation and schematic of nonspecific background adhesion of cells, and the calculated f* as a function of RGD nanospacing normalized to f* at 67 nm nanospacing.
Finally, combining author’s previous work with the work of other scientists, the author provides an explanation of the minimum effective ligand nanospacing at the level of biomacromolecules and cytoskeleton networks, and summarize the relationship between RGD nanospacing and cell adhesion for the first time (Figure 8). The basic theoretical research of Ding's group provides a more comprehensive understanding of cell-material interactions at the micron and nanometer scales.
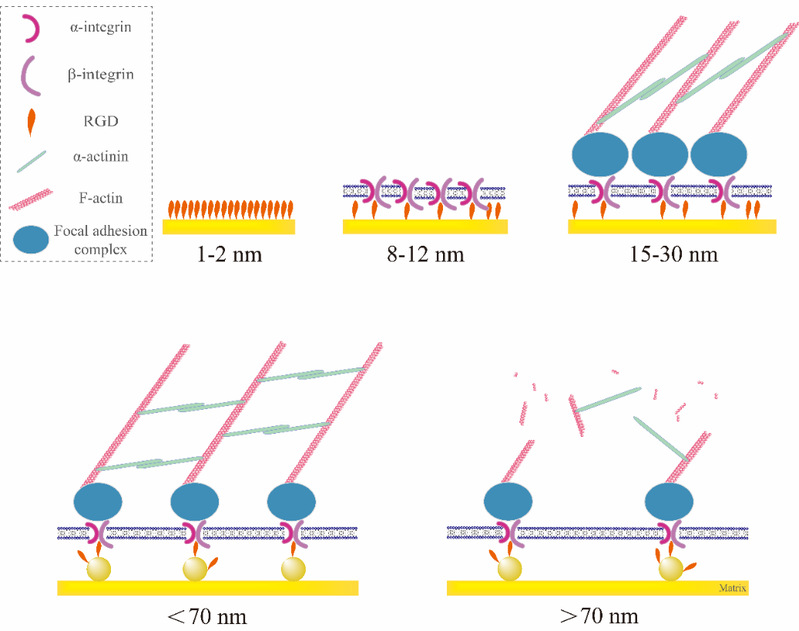
Figure 8 Schematic illustration showing the underlying RGD nanospacings to regulate cell adhesion.Upper row: Even if the RGD peptide is densely grafted onto gold surfaces, the steric hindrance of the RGD peptide size will result in a spacing of 1 - 2 nm between the nearest two RGDs. After one-to-one bioconjugating between an integrin and the ligand(s) at one small nanodot, some RGDs grafted on gold nanodots might be redundant for a static adhesion. So, the minimal effective nanospacing follows the integrin size of 8 - 12 nm. Integrins connect FA complex inside the membrane and interacts with cytoskeletal components to trigger focal adhesion. Combination of the dimension of a FA complex, the feature size along the material surface might be about 15 to 30 nm, which primarily interprets the minimal effective RGD nanospacing for an unpatterned gold surface with densely grafted RGDs, namely about 19 nm as shown in Figure 5. Lower row: When the RGD nanopacing is less than 70 nm, the cells can form a complete and powerful cytoskeleton junction, which is reflected in the macroscopic good adhesion of cells. When the RGD nanospacing is larger than 70 nm, due to the limitation of α-actinin length, stable cytoskeleton structure cannot be formed inside the cell, which is reflected in the significantly worse cell adhesion.
The article was published in the journal of Nano Researchwith Ms. Shuang Zheng as first author and Professor Jiandong Ding as corresponding author. See please: Shuang Zheng, Qiong Liu, Junhao He, Xinlei Wang, Kai Ye, Xuan Wang, Ce Yan, Peng Liu, Jiandong Ding*, Critical adhesion areas of cells on micro-nanopatterns, Nano Research (2021), doi.org/10.1007/s12274-021-3711-6
Get to know us better now!

Wechat:FDUMMers
Search!
Search across our website
Revenant @ 2018 by fudan | All Rights Reserved
Powered by Weicheng

